Survival analysis
https://web.stanford.edu/~lutian/coursepdf/
Log-rank test with a simple example
Let’s start with a simple example. Suppose we have Group 1 and Group 2 and we want to test whether the two groups have the same survival function or not.
Group 1:
| time | status |
|---|---|
| 2 | 1 |
| 3 | 1 |
| 5 | 0 |
| 7 | 1 |
| 7 | 1 |
| 8 | 1 |
Group 2:
| time | status |
|---|---|
| 2 | 1 |
| 2 | 1 |
| 4 | 1 |
| 4 | 1 |
| 6 | 1 |
| 8 | 1 |
In the status, 1 means an event, and 0 means a censored.
There were 6 different time points where events (or censored) were recorded. They are {2, 3, 4, 6, 7, 8}. We do another bookkeeping as following.
Group1 Group2
| time | | | | | | |
|---|---|---|---|---|---|---|
| 2 | 1 | 0 | 6 | 2 | 0 | 6 |
| 3 | 1 | 0 | 5 | 0 | 0 | 4 |
| 4 | 0 | 1 | 4 | 2 | 0 | 4 |
| 6 | 0 | 0 | 3 | 1 | 0 | 2 |
| 7 | 2 | 0 | 3 | 0 | 0 | 1 |
| 8 | 1 | 0 | 1 | 1 | 0 | 1 |
counts events and counts censored. is for at-risk counts. The interpretation for the first row is as follows: before the time point 2, there were 6 at-risk both in Group 1 and Group 2. There were 1 event in Group 1 and 2 events in Group 2. None was censored in both groups. Likewise, the interpretation of the third row is as follows: there were four individuals at risk in both Group 1 and Group 2. At time point 4, one individual in Group 1 was censored, and two events occurred in Group 2. However, if we refer to the status table above, we see that the censoring actually occurred at time point 5, not 4. Although censored individuals are not counted as events, we include them in the risk set at the immediately preceding time point, rather than at the time they were censored.
If two groups follow the same hazard function, we will estimate that the hazard rate at time 2 is 3/12. It is So, we calculate the expected number of events by
| time | | | | | | |
|---|---|---|---|---|---|---|
| 2 | 1 | 6 | 2 | 6 | 1.5 | 1.5 |
| 3 | 1 | 5 | 0 | 4 | 0.56 | 0.44 |
| 4 | 0 | 4 | 2 | 4 | 1 | 1 |
| 6 | 0 | 3 | 1 | 2 | 0.6 | 0.4 |
| 7 | 2 | 3 | 0 | 1 | 1.5 | 0.5 |
| 8 | 1 | 1 | 1 | 1 | 1 | 1 |
Note that .
We also see that if the hazard rates are same, the difference in of both groups is just a matter of random split. That is, follows , hypergeometric distribution with the population size of , the success count of , and the draw size of .
So the expected value of is
The variance of is
By the Lyapunov central limit theorem,
where
In fact, there is a symmetry in , so .
| time | | | | | | | |
|---|---|---|---|---|---|---|---|
| 2 | 1 | 6 | 2 | 6 | 0.613 | 1.5 | -0.5 |
| 3 | 1 | 5 | 0 | 4 | 0.246 | 0.56 | 0.44 |
| 4 | 0 | 4 | 2 | 4 | 0.428 | 1 | -1 |
| 6 | 0 | 3 | 1 | 2 | 0.24 | 0.6 | -0.6 |
| 7 | 2 | 3 | 0 | 1 | 0.25 | 1.5 | 0.5 |
| 8 | 1 | 1 | 1 | 1 | 0 | 1 | 0 |
So, and .
. So, we won’t be able to reject the null hypothesis.
By the way, follows .
Let’s see no-roundup calculation using R code.
df <- data.frame(m1=c(1, 1, 0, 0, 2, 1))
df$n1 <- c(6,5,4,3,3,1)
df$m2 <- c(2,0,2,1,0,1)
df$n2 <- c(6,4,4,2,1,1)
df$E <- df$n1*(df$m1 + df$m2)/(df$n1 + df$n2)
df$V <- ( df$n1*df$n2 )/( (df$n1 + df$n2)^2 )*( df$m1 + df$m2 )*(df$n1 + df$n2 -df$m1-df$m2 )/(df$n1 + df$n2 - 1 )
s_square = sum(df$V)
OmE_square = (sum(df$m1 - df$E))^2
z = sqrt(OmE_square/s_square)
chisq_stat = OmE_square/s_square
cat('z =',z)
cat('\nchisq =', chisq_stat)
cat('\np_val_z = ', 2*(1-pnorm(z)))
cat('\np_val_chisq = ', (1-pchisq(chisq_stat, df=1)))z = 0.8663394
chisq = 0.7505439
p_val_z = 0.3863041
p_val_chisq = 0.3863041If we use R libraries, we can see familiar numbers.
R code chunks
💡Libraries
library(survival) library(survminer)💡Create dataset
# 'time' is the observed time until event or censoring # 'event' is the event indicator (1 = event occurred, 0 = censored) # 'batch' is a categorical variable for manufacturing batch simple_data <- data.frame( time = c(2, 3, 5, 7, 7, 8, # Fertilizer X 2, 2, 4, 4, 6, 8), # Fertilizer Y event = c(1, 1, 0, 1, 1, 1, # Fertilizer X events/censoring 1, 1, 1, 1, 1, 1), # Fertilizer Y events/censoring group = factor(c(rep("Group 1", 6), rep("Group 2", 6))) # Create factors for groups )💡Create a 'Surv' object
# This object combines the time and event status, which is required by # survival analysis functions in R. surv_object_simple <- Surv(time = simple_data$time, event = simple_data$event)💡Kaplan-Meier estimation for overall survival
km_fit_simple <- survfit(surv_object_simple ~ group, data = simple_data) # Print the summary of the overall KM fit print("Summary of Kaplan-Meier Fit by Group 1 or 2:") print(summary(km_fit_simple))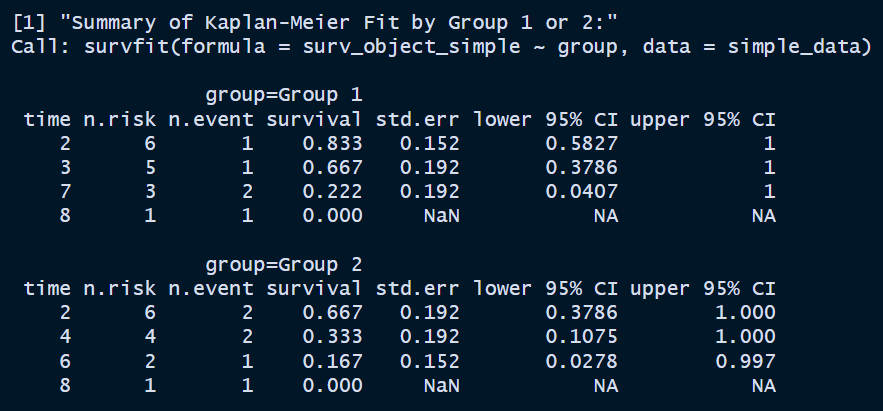
Here, the
survivalcolumn is the estimated survival function values. The formula is . The first value of Group 1 is . The second value is . Thestd.errcolumn is calculated by Greenwood’s formula, which is . The95% CIcalculation has a few options:plain,log, orlog-log. It appears thatlogtransformation based method is applied here. The formula isFor the first row of Group 1,
sounds about right. For the second row
also looks correct enough. Which one to use when? Let’s ask chatGPT.

It sounds like
survfitchose something that’s rarely used nowadays.💡5. Plot the Kaplan-Meier curves by group
# ggsurvplot from survminer provides a nice, publication-ready plot ggsurvplot(km_fit_plant, data = plant_data, risk.table = TRUE, linetype = c(1,4), tables.height = 0.3, pval = TRUE)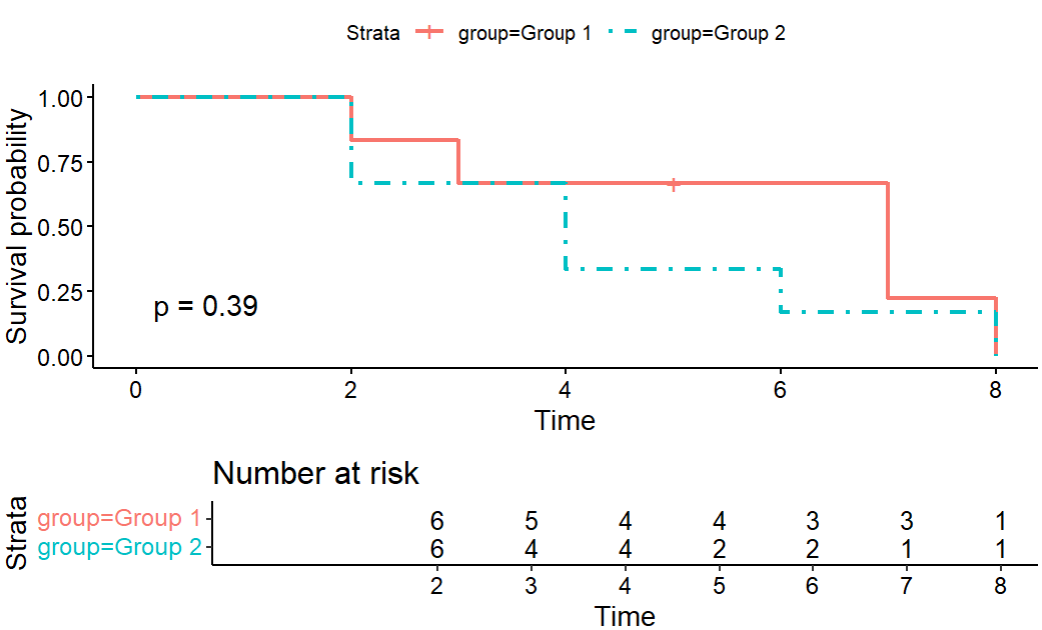
More to add with a complex example
Code chunks
#https://bookdown.org/drki_musa/dataanalysis/survival-analysis-kaplan-meier-and-cox-proportional-hazard-ph-regression.html#data library(here) library(tidyverse) library(lubridate) library(survival) library(survminer) library(broom) library(gtsummary)# https://github.com/drkamarul/data-for-RMed/tree/main/data stroke <- read_csv(here('.', 'stroke_data.csv')) KM <- survfit(Surv(time = time2, event = status == "dead" ) ~ 1, data = stroke) summary(KM)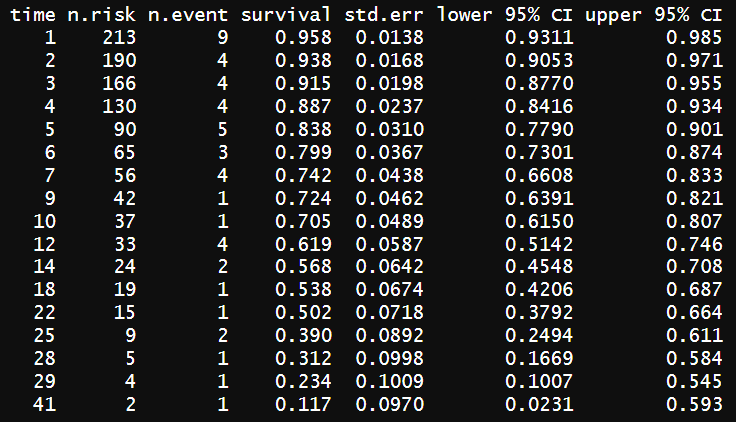
Let try the CI calculation again. The first row:
looks good.
# for stoke types KM_str_type2 <- survfit(Surv(time = time2, event = status == "dead" ) ~ stroke_type, data = stroke) summary(KM_str_type2)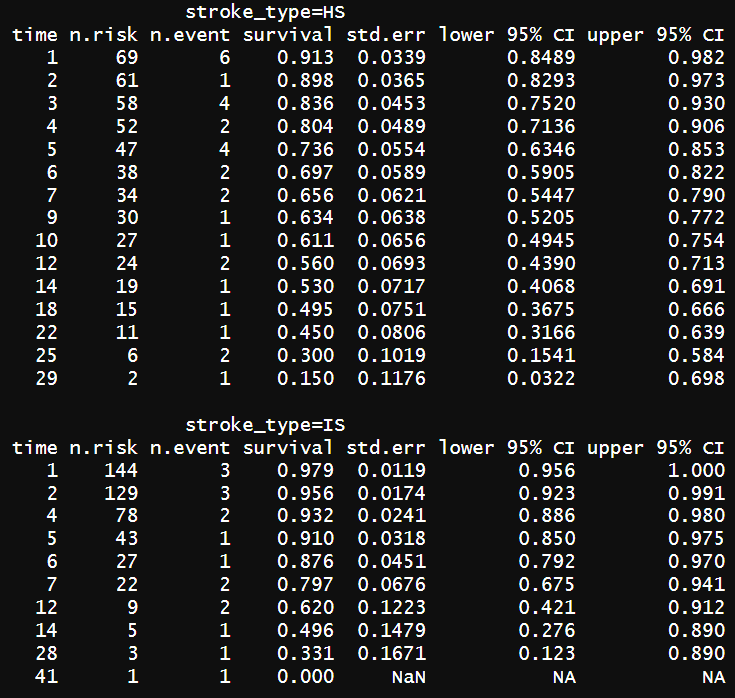
ggsurvplot(KM_str_type2, data = stroke, risk.table = TRUE, linetype = c(1,4), tables.height = 0.3, pval = TRUE)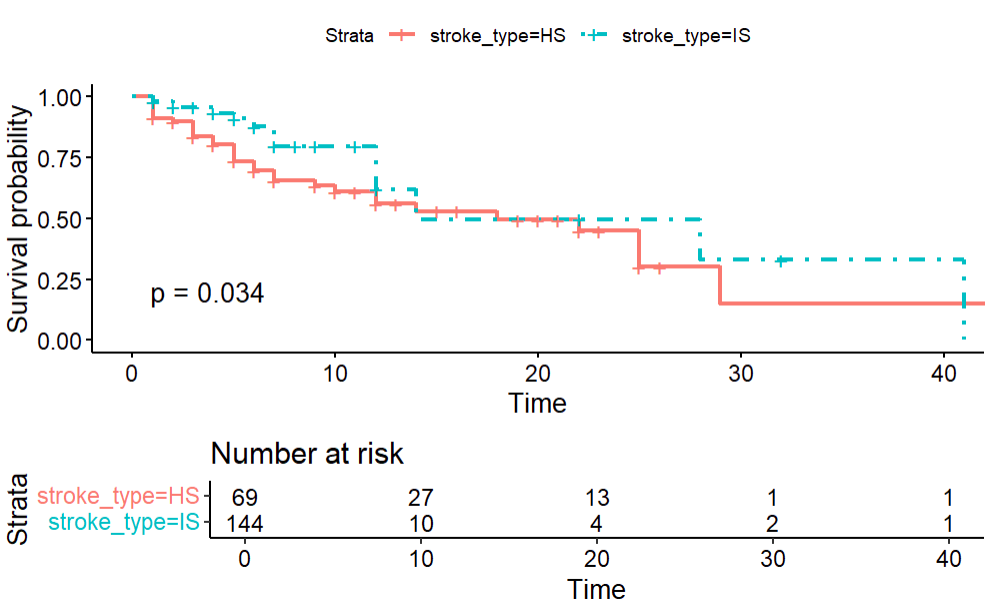
survival functions are different when groups are divided by stroke types.
#For diebete KM_dm <- survfit(Surv(time = time2, event = status == "dead" ) ~ dm, data = stroke) summary(KM_dm)ggsurvplot(KM_dm, data = stroke, risk.table = TRUE, linetype = c(1,4), tables.height = 0.3, pval = TRUE)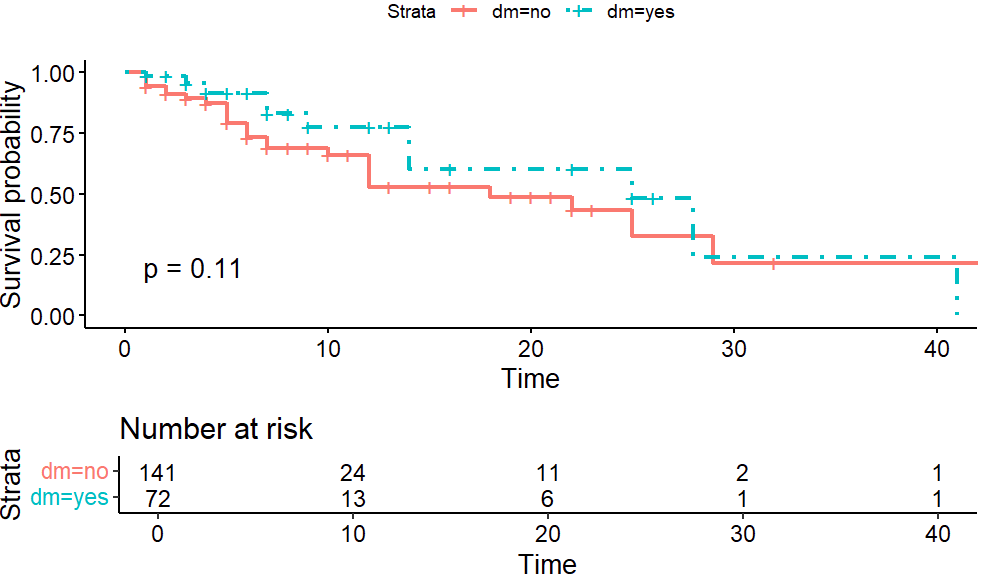
survival functions are not significantly different when groups are divided by dm.
Log-rank test. (It’s been done already)
survdiff(Surv(time = time2, event = status == "dead") ~ stroke_type, data = stroke, rho = 0) ``` Call: survdiff(formula = Surv(time = time2, event = status == "dead") ~ stroke_type, data = stroke, rho = 0) N Observed Expected (O-E)^2/E (O-E)^2/V stroke_type=HS 69 31 24.2 1.92 4.51 stroke_type=IS 144 17 23.8 1.95 4.51 Chisq= 4.5 on 1 degrees of freedom, p= 0.03survdiff(Surv(time = time2, event = status == "dead") ~ dm, data = stroke, rho = 0) ## Call: survdiff(formula = Surv(time = time2, event = status == "dead") ~ dm, data = stroke, rho = 0) N Observed Expected (O-E)^2/E (O-E)^2/V dm=no 141 35 29.8 0.919 2.54 dm=yes 72 13 18.2 1.500 2.54 Chisq= 2.5 on 1 degrees of freedom, p= 0.1What is peto-peto test? Let’s see what we get.
survdiff(Surv(time = time2, event = status == "dead") ~ stroke_type, data = stroke, rho = 1) ## Call: survdiff(formula = Surv(time = time2, event = status == "dead") ~ stroke_type, data = stroke, rho = 1) N Observed Expected (O-E)^2/E (O-E)^2/V stroke_type=HS 69 25.3 18.7 2.33 6.02 stroke_type=IS 144 13.7 20.3 2.15 6.02 Chisq= 6 on 1 degrees of freedom, p= 0.01Hmm, p-value is lower. Nice!
survdiff(Surv(time = time2, event = status == "dead") ~ dm, data = stroke, rho = 1) ## Call: survdiff(formula = Surv(time = time2, event = status == "dead") ~ dm, data = stroke, rho = 1) N Observed Expected (O-E)^2/E (O-E)^2/V dm=no 141 29.54 24.3 1.11 3.61 dm=yes 72 9.41 14.6 1.85 3.61 Chisq= 3.6 on 1 degrees of freedom, p= 0.06According to the chatGPT,
- Peto-Peto (Modified Wilcoxon) Test
- test for differences between survival curves, but gives more weight to earlier time points
- test for differences between survival curves, but gives more weight to earlier time points
- Peto-Peto (Modified Wilcoxon) Test
Dig-ressions
Cox proportional hazards regression
The hazard function is a rate, and because of that it must be strictly positive.
One form of a regression model for the hazard function that addresses the study goal is:
Here, is variables like certain medical conditions and is a vector of parameters.
- The function, , characterizes how the hazard function changes as a function of survival time. baseline hazard function.
- The function, , characterizes how the hazard function changes as a function of subject covariates, like medical conditions.
The hazard ratio (HR) depends only on the function .
If the model hazard function is
the hazard ratio is
This model is referred to in the literature by a variety of terms, such as the Cox model, the Cox proportional hazards model or simply the proportional hazards model.
So for example, if we have a covariate which is a dichomotomous (binary), such as stroke type: coded as a value of and , for HS and IS, respectively, then the hazard ratio becomes
If the value of the coefficient is , then the interpretation is that HS are dying at twice () the rate of patients with IS.
Some disadvantages of KM
- It is a univariable analysis
- It is a non-parametric analysis
We also acknowledge that the fully parametric regression models in survival analysis have stringent assumptions and distribution
requirement. So, to overcome the limitations of the KM analysis and the fully parametric analysis, we can model our survival data using the
semi-parametric Cox proportional hazards regression.
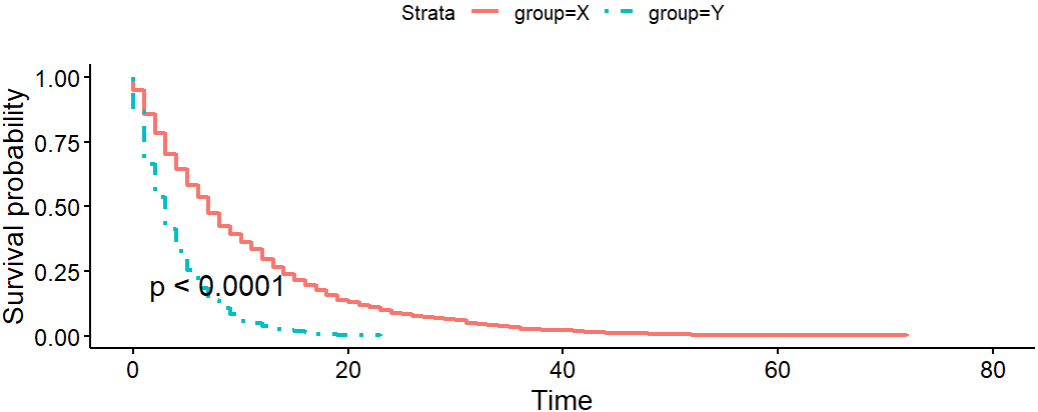
Let’s apply this to the data generated in ⛏️Digression - simulate survival function.
The hazard ratio is 2.75. .
sim_data_coxph <-
coxph(Surv(time = time,
event = event == 1) ~ group,
data = simulated_data)
summary(sim_data_coxph)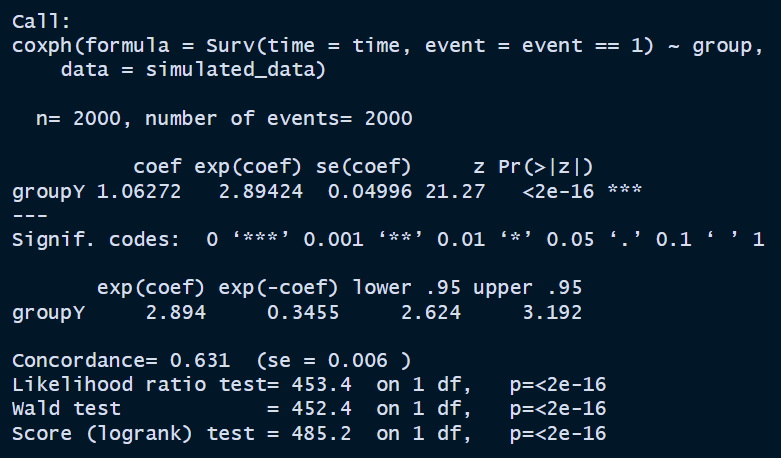
It looks like exp(coef) is very close to 2.75.
Estimation from Cox proportional hazards regression
Simple Cox PH regression
Using our stroke dataset, we will estimate the parameters using the Cox PH regression. Remember, in our data we have
- the time variable :
time2
- the event variable :
statusand the event of interest is dead. Event classified other than dead are considered as censored.
- date variables : date of admission (doa) and date of discharge (dod)
- all other covariates
stroke_stype <-
coxph(Surv(time = time2,
event = status == 'dead') ~ stroke_type,
data = stroke)
summary(stroke_stype)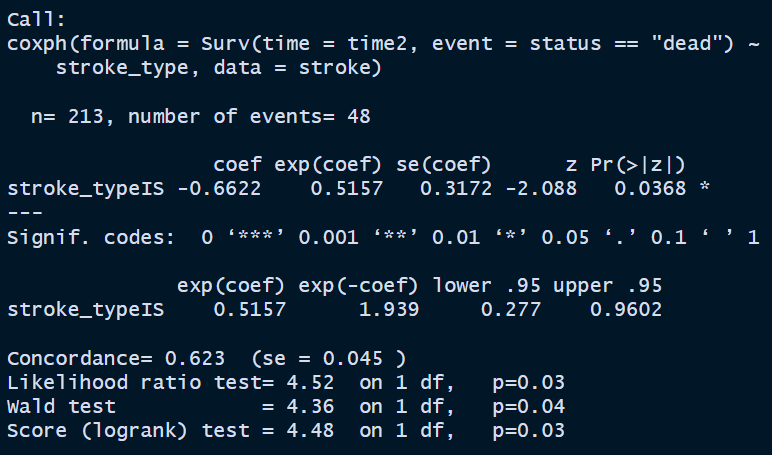
tidy(stroke_stype,
conf.int = TRUE)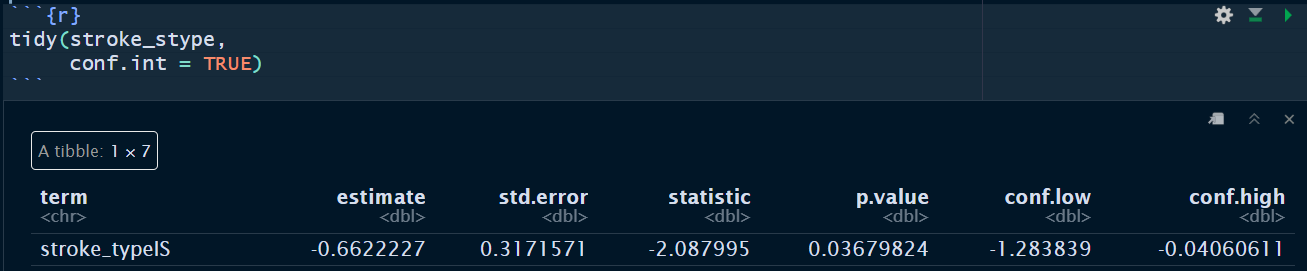
The simple Cox PH model with covariate stroke type shows that the patients with IS has −0.0662 times the crude log hazard for death as compared to patients with HS (p-value = 0.0368).
This means that
and
So, patients with IS survived more.