Digression - simulate survival function
: Probability of “death” by time t. means that half will die by time 100.
is a cumulative distribution of , with .
: Survival function, , probability to survive to time t.
: hazard function. the instantaneous failure rate, scaled by the fraction of surviving systems at time t
This means that
Or,
Let’s simulate a survival data for a simple case.
Let’s suppose that we have a survival data whose hazard function is constant, .
Since and is a monotone decreasing function, a constant hazard function means that , the probability density function of the even happening, should be also a monotone decreasing function.
By the above formula, . We will generate event occurring time data by inverse transformation sampling. That means, , for u from uniform(), is a random sample. We will be solving . By the way, is also a random number from uniform(). So, we will just solve for from . That is, .
set.seed(5670)
h = 0.1
n <- 20
u <- runif(n)
observed_times <- -log(u)/h
status <- as.numeric(observed_times >-1) # let's assume that non was censored.
surv_obj <- Surv(round(observed_times), status) # observed times are rounded as if the unit of time is one.
# Fit Kaplan-Meier estimator
fit <- survfit(surv_obj ~ 1)
# Plot survival curve
plot(fit, xlab = "Time", ylab = "Survival Probability", main = "Simulated Survival Curve")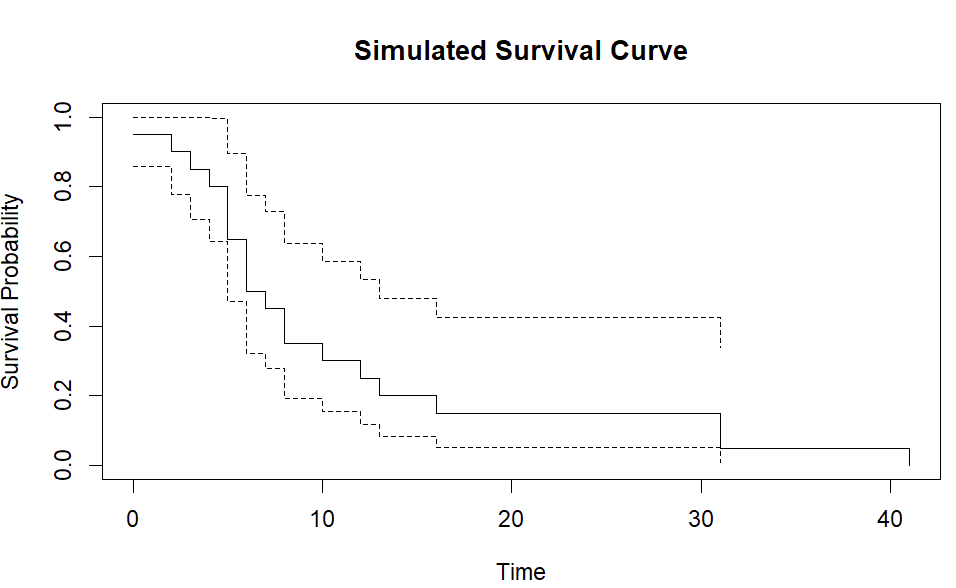
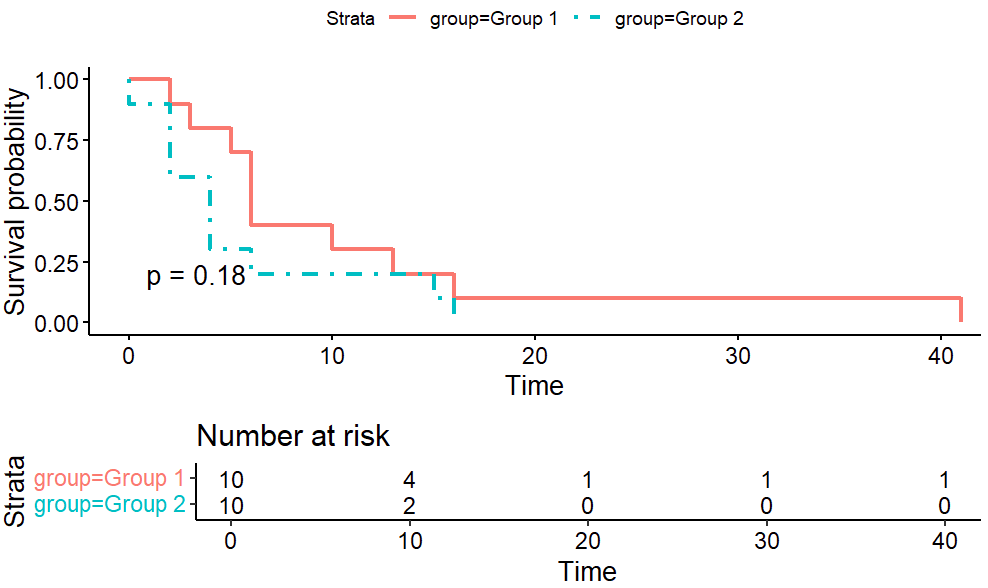
The following is a simulation of two groups one is with and the other has . There are 10 subjects in each group. With a total of 20 subjects, the p-value of log-rank test is 0.18.
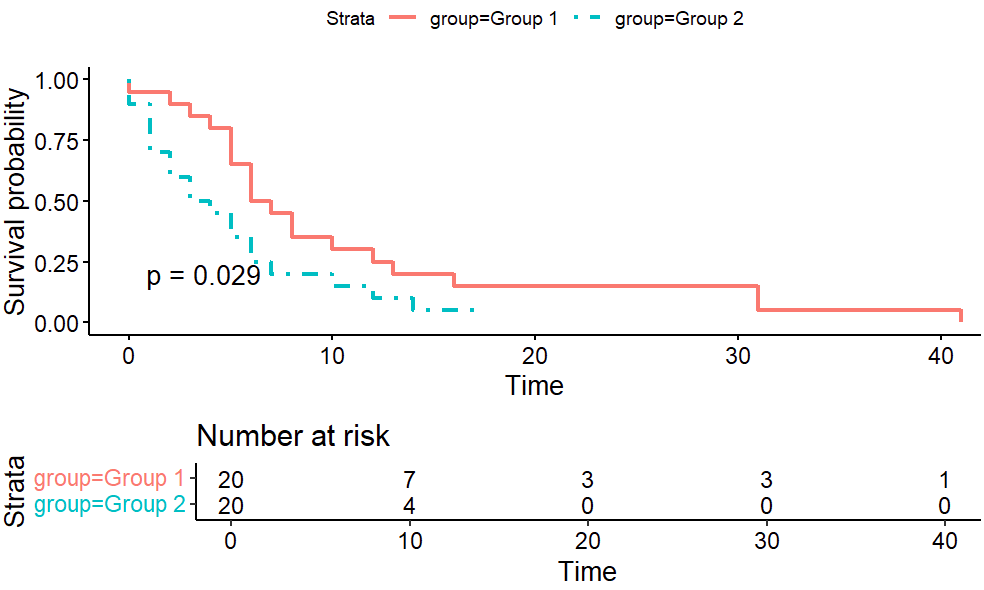
Here we increase the number of subjects to 20 each.
2000 simulated data
Let’s generate 2*1000 simulated data for the Cox proportional hazard regression. Two groups, X and Y have a constant hazard function ratio of 2.75.
# set.seed(5670)
n <- 1000
h = 0.1
u1 <- runif(n)
observed_times1 <- -log(u1)/h
h2 = h*2.75
u2 <- runif(n)
observed_times2 <- -log(u2)/h2
# status <- as.numeric(observed_times >-1)
simulated_data <- data.frame(
time = round(c(observed_times1, observed_times2)), # Fertilizer Y
event = rep(1, 2*n), # Fertilizer Y events/censoring
group = factor(c(rep("X", n), rep("Y", n))) # Create factors for groups
)
surv_object_simulated <- Surv(time = simulated_data$time, event = simulated_data$event)
# surv_obj <- Surv(round(observed_times), status)
km_fit_simulated <- survfit(surv_object_simulated ~ group, data = simulated_data)
# Print the summary of the overall KM fit
print("Summary of Kaplan-Meier Fit by Group 1 or 2:")
print(summary(km_fit_simulated))