graphRAG on Rivanna (HPC) - demo
The motivation to consider a graphRAG for biological findings is 1. The collection of biological findings can be repetitive as sentences collection. 2. There can be associated numerical data to each findings. Both seem to make it difficult for LLM learn about biological findingsas texts.
Graph DB building
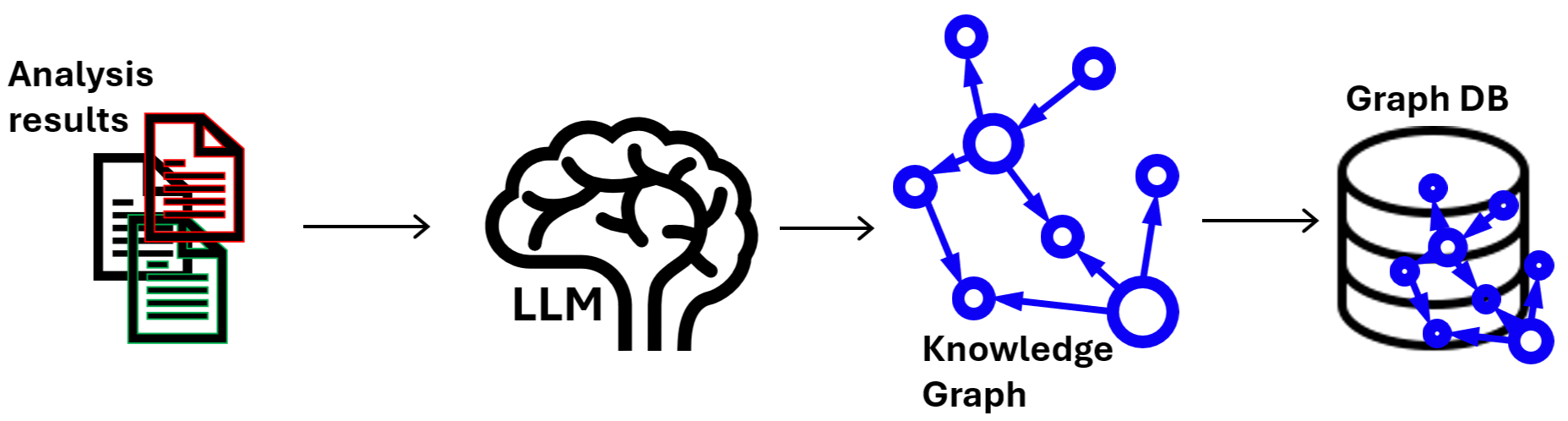
graphRAG structure
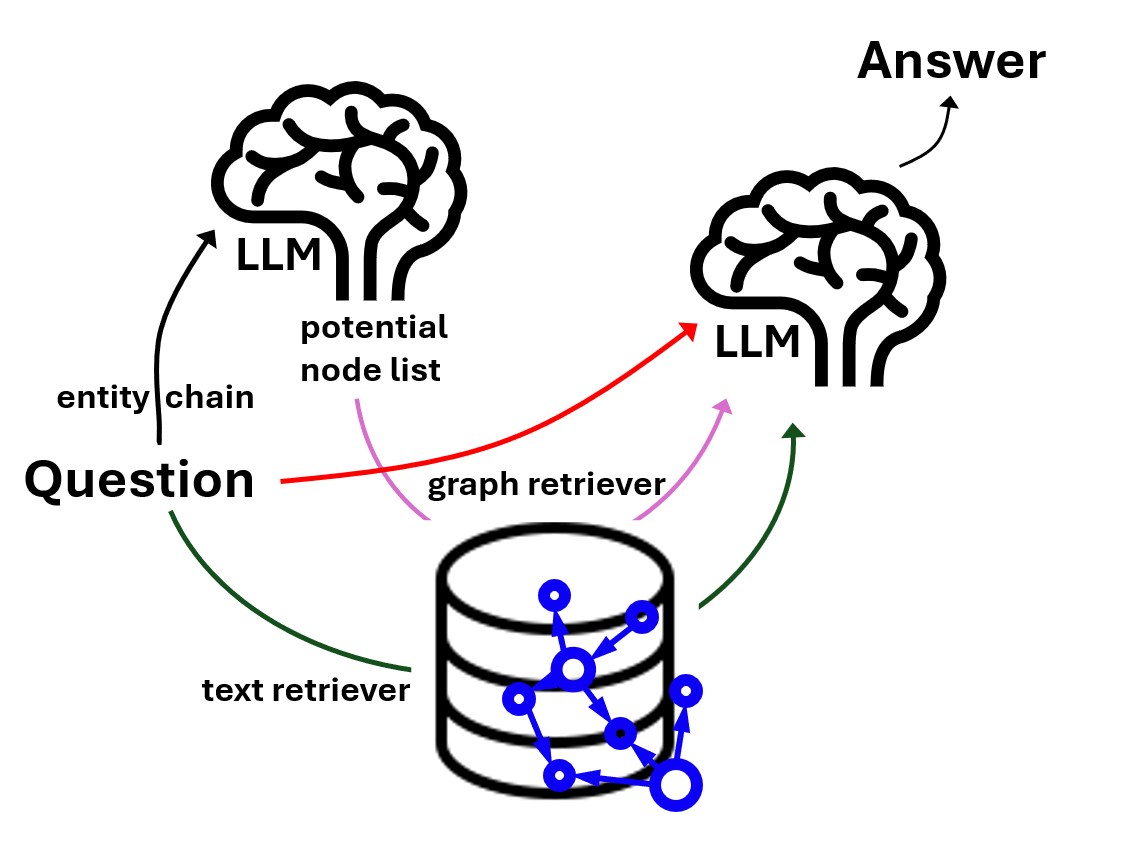
graphRAG with a bit of extra features
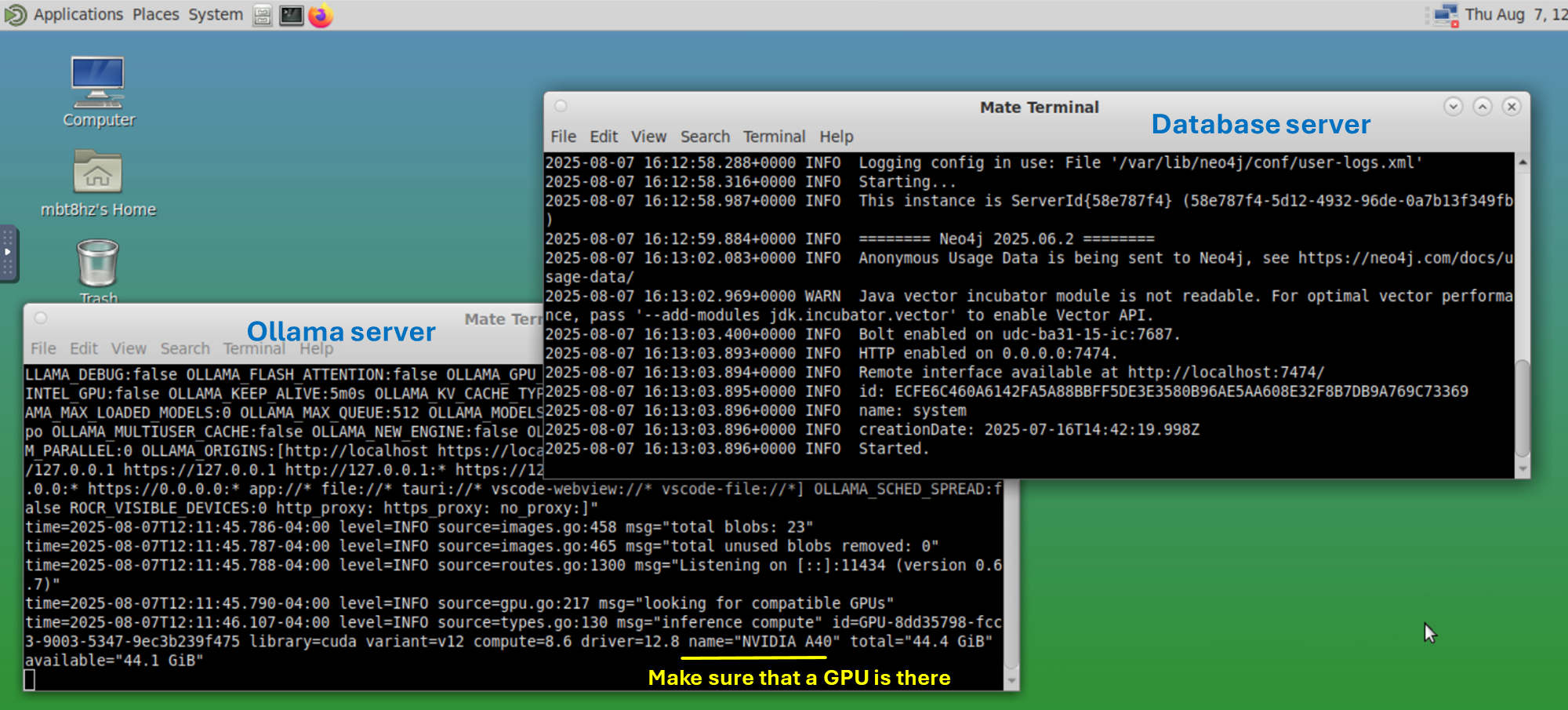
Local LLM Database servers for graphRAG
With Ollama server and graph database server running
Feed the following text to LLM (I am using Phi4 here)
text = """
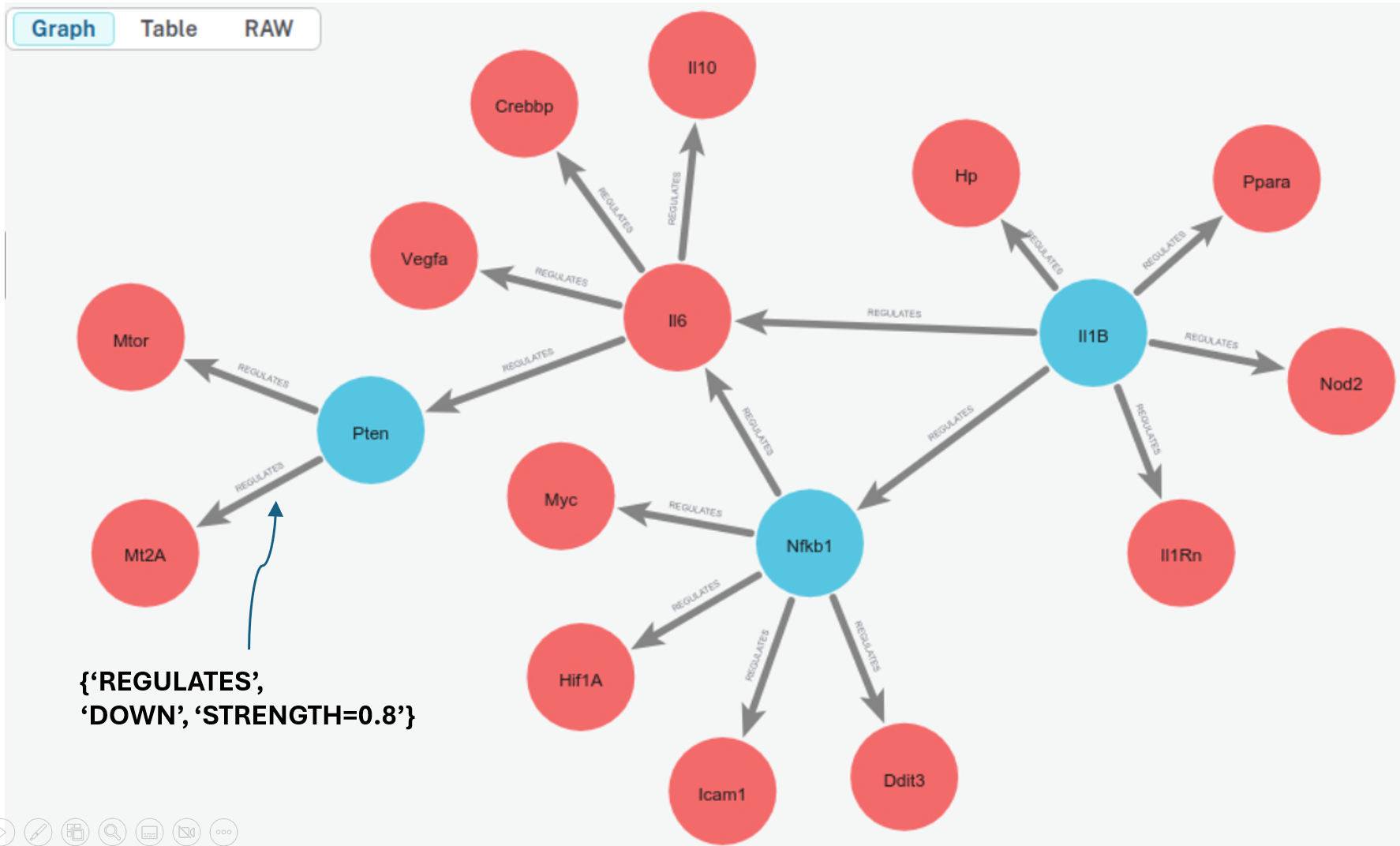
The transcription factor IL1B regulates the HP gene.
The transcription factor IL1B regulates the IL1RN gene.
The transcription factor IL1B regulates the IL6 gene.
The transcription factor IL1B regulates the NOD2 gene.
The transcription factor IL1B regulates the NFKB1 gene.
The transcription factor IL1B regulates the PPARA gene.
The transcription factor IL6 regulates the IL10 gene.
The transcription factor IL6 regulates the CREBBP gene.
The transcription factor IL6 regulates the PTEN gene.
The transcription factor IL6 regulates the VEGFA gene.
The transcription factor NFKB1 regulates the MYC gene.
The transcription factor NFKB1 regulates the HIF1A gene.
The transcription factor NFKB1 regulates the DDIT3 gene.
The transcription factor NFKB1 regulates the ICAM1 gene.
The transcription factor NFKB1 regulates the IL6 gene.
The transcription factor PTEN regulates the MT2A gene.
The transcription factor PTEN regulates the MTOR gene.
"""The LLM employed by LangChain will read the text and turn into knowledge graphs. Save the graph on the database.
Let’s see how smart this can be.
💡
LLM interface, smart interface
A few internal components

Questions/Tasks
ans_0 = chain.invoke(input="Based on the provided document, does NFKB1 regulate AHR?")
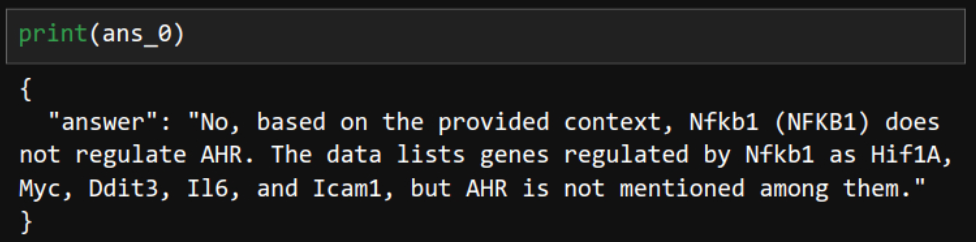
Correct. It was an easy question.
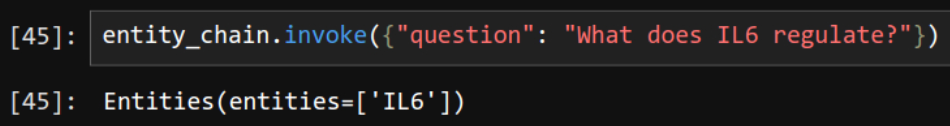
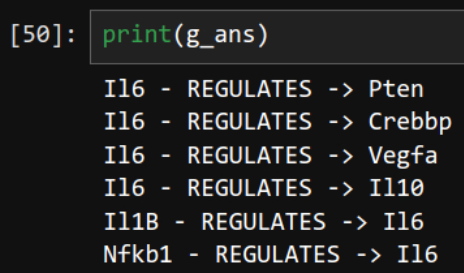
ans_100 = chain.invoke(input="Find all genes that transctiption factor IL6 regulates. Give me one short sentence answer.")
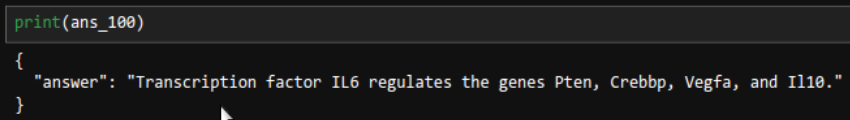
Correct. It was an easy question, too.
# 2-hop neighbors
ans_200 = chain.invoke(input="If A regulates B and B regulates C, then A also regulates C. Considering this and the provided document, find all genes that IL1B regulates.")
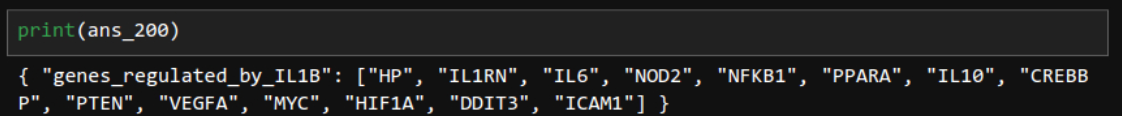
Correct. LLM understood transitivity. Smart!!!
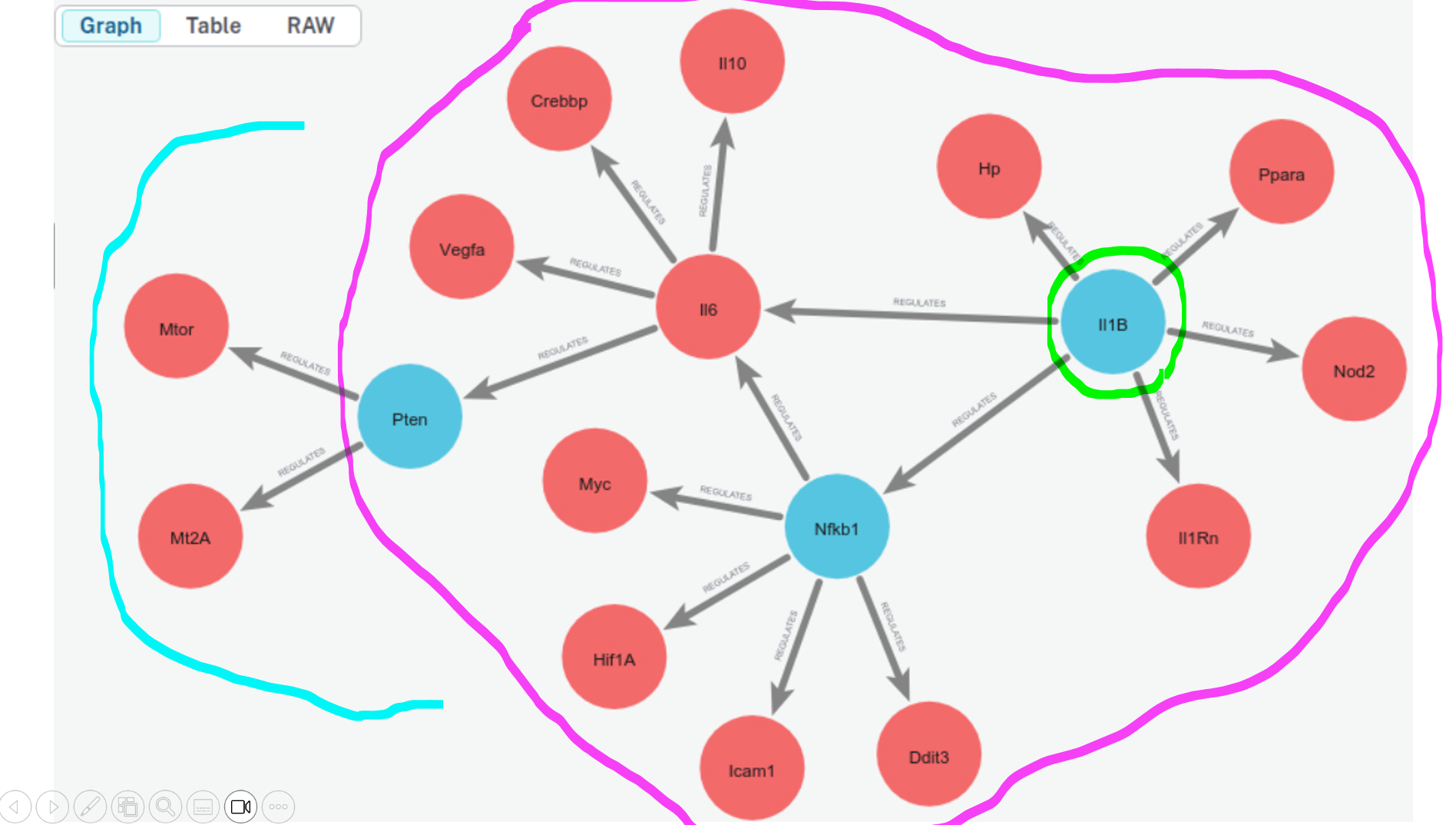
ans_300 = chain.invoke(input="If gene A regulates gene B, and gene B regulates gene C, then gene A is also considered to regulate gene C. Based on this transitive relationship and the provided document, identify all genes regulated by IL1B. Write one sentence for the answer.")
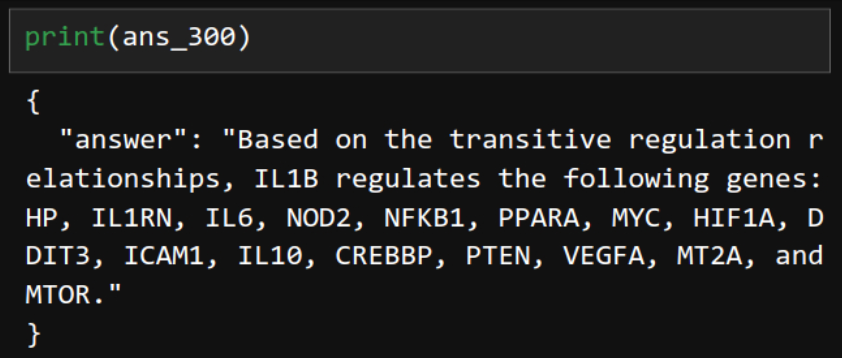
Correct. Transitivity all the way.
ans_400 = chain.invoke(input="Based on up to 2-hop neighbors in the provided document, identify everything regulated by IL1B. Write one sentence for the answer.")
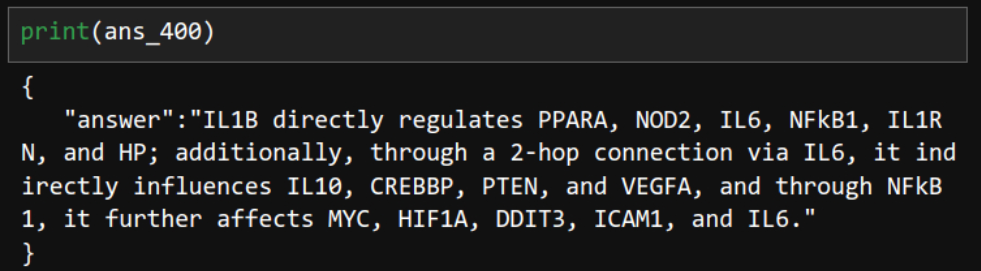
Correct — it also recognized 2-hop neighbors. “Sorry for underestimating you!” However, there's a caveat: in this task, the word "everything" played a key role. Without it, the LLM's response was slightly off.
Tapping on edge properties.
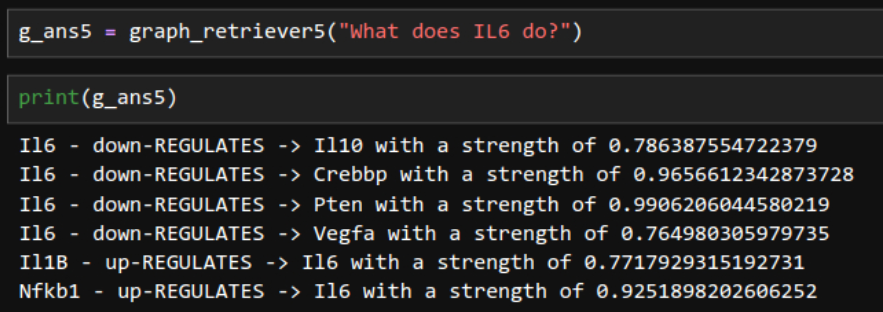
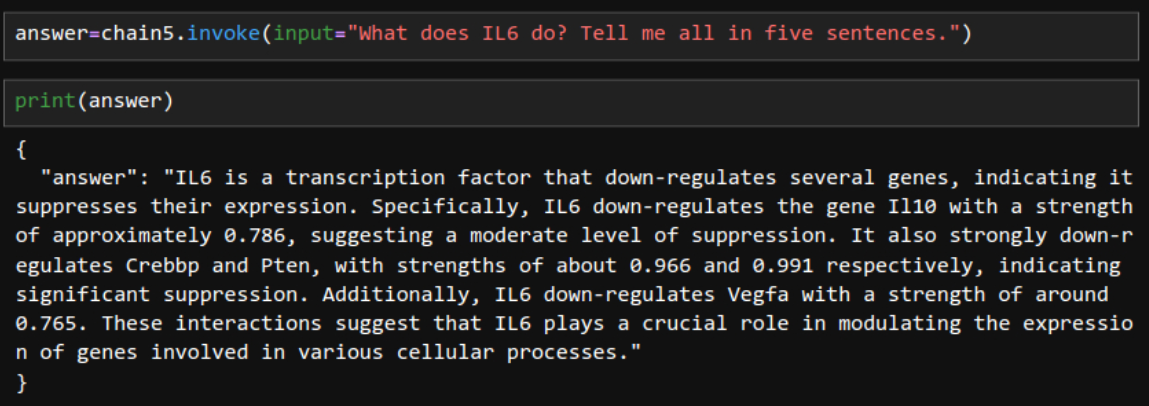
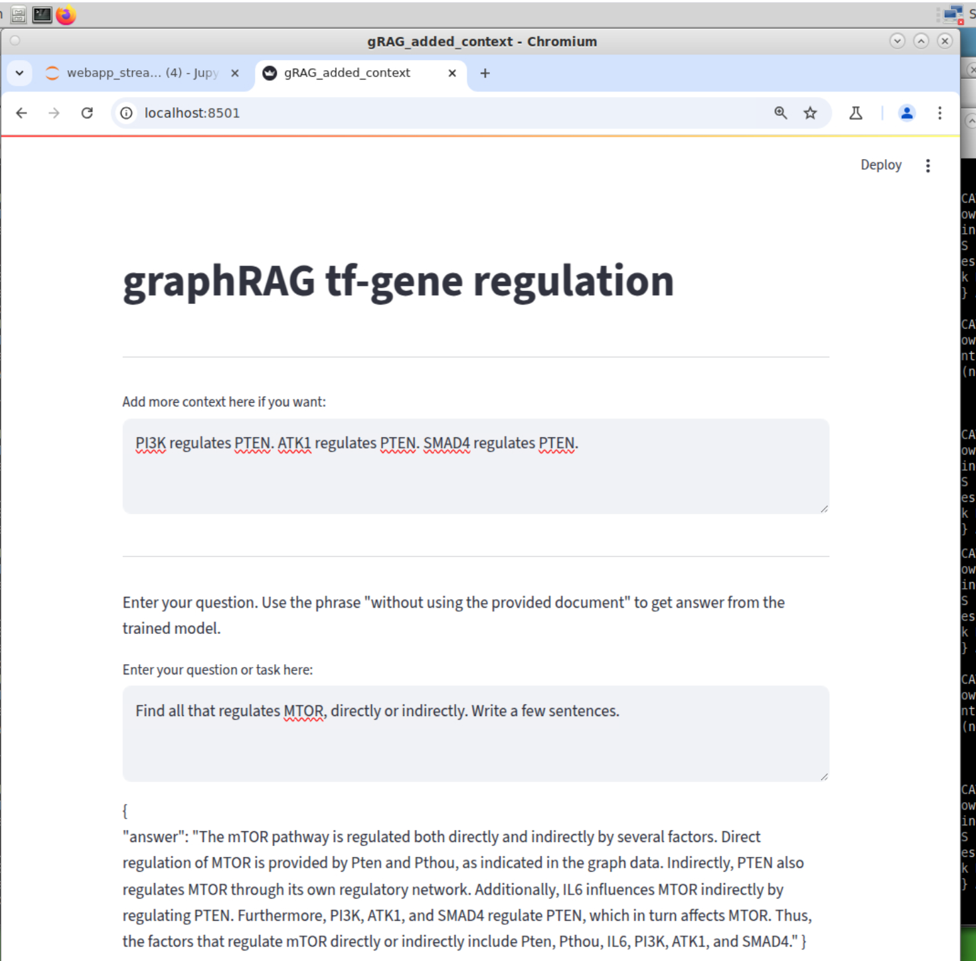
Web Interface. Now it looks like a web-based LLM model.
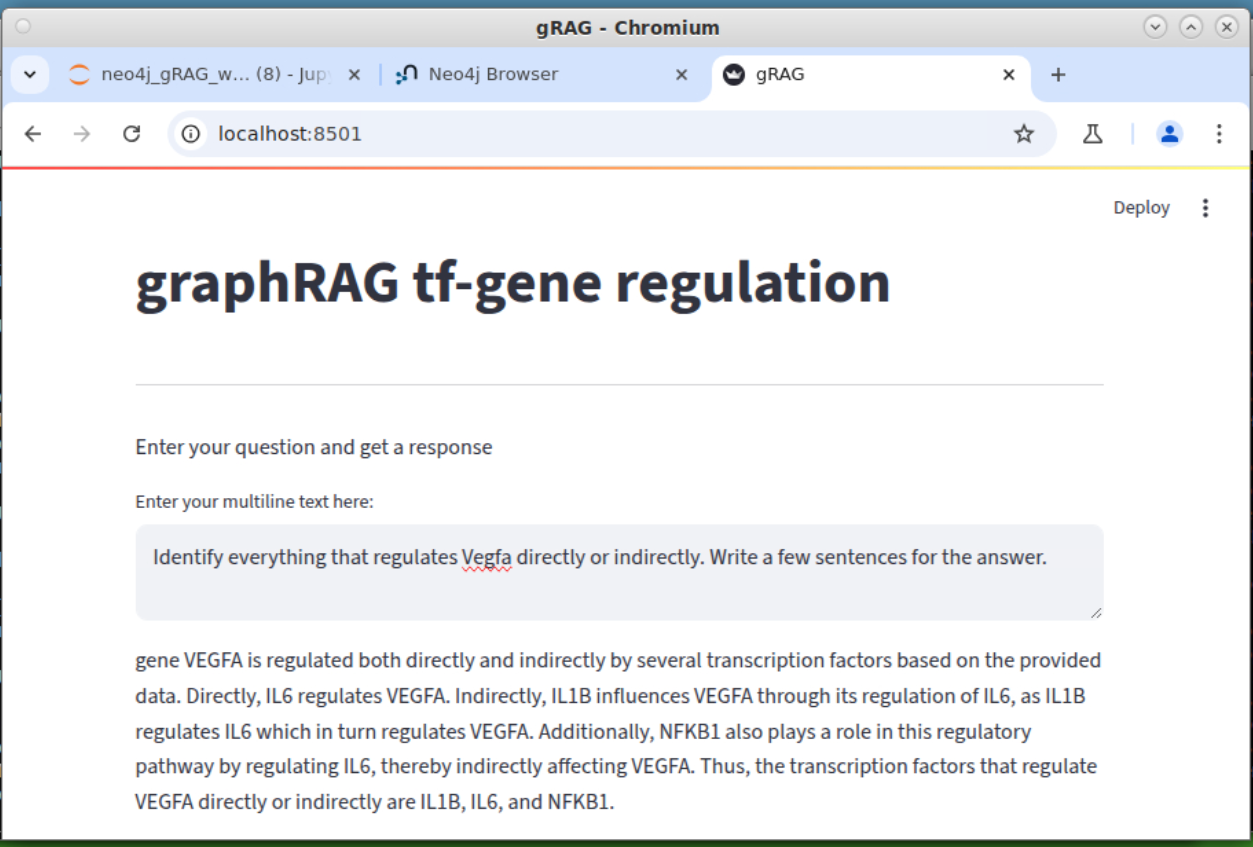
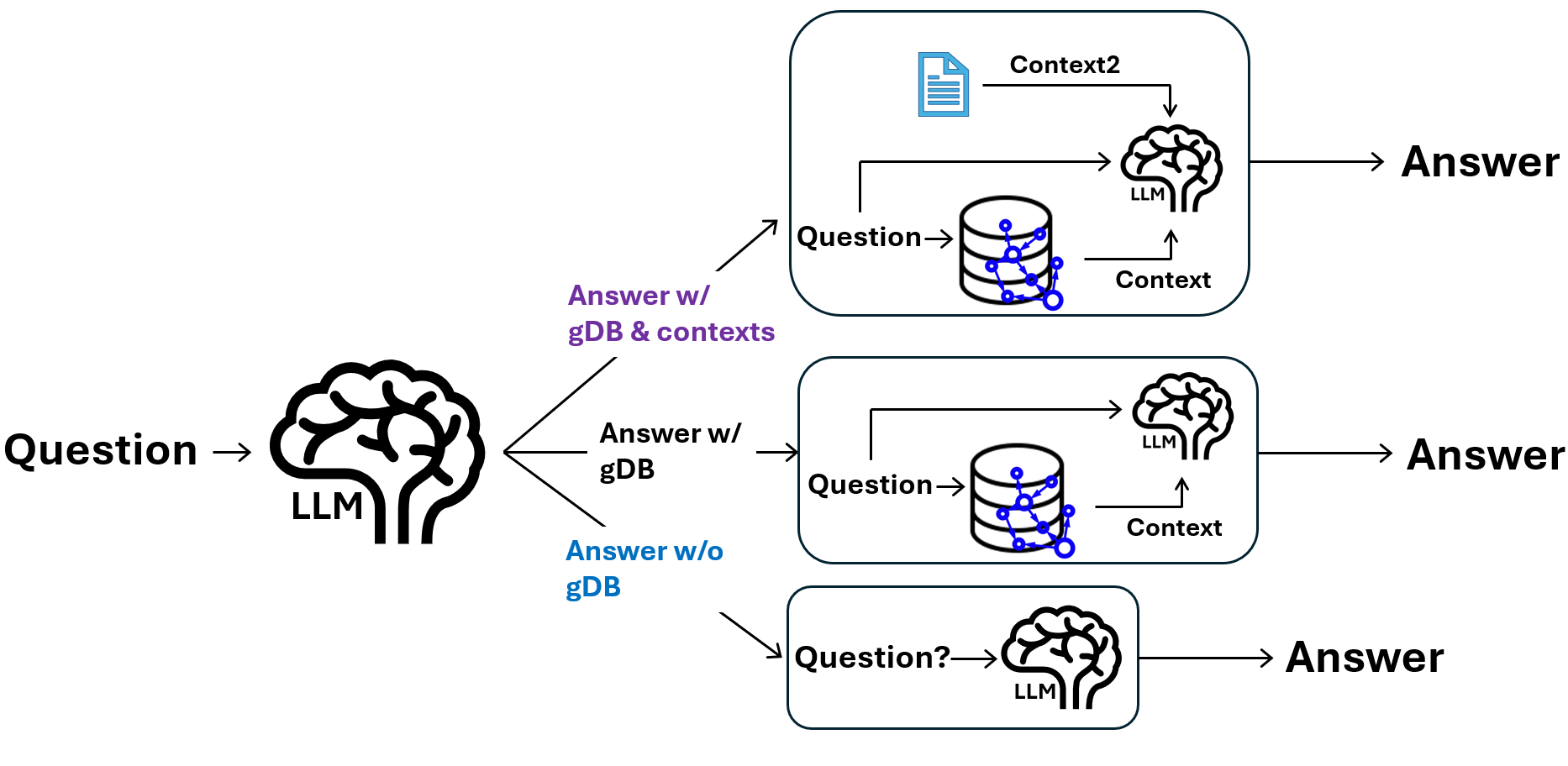
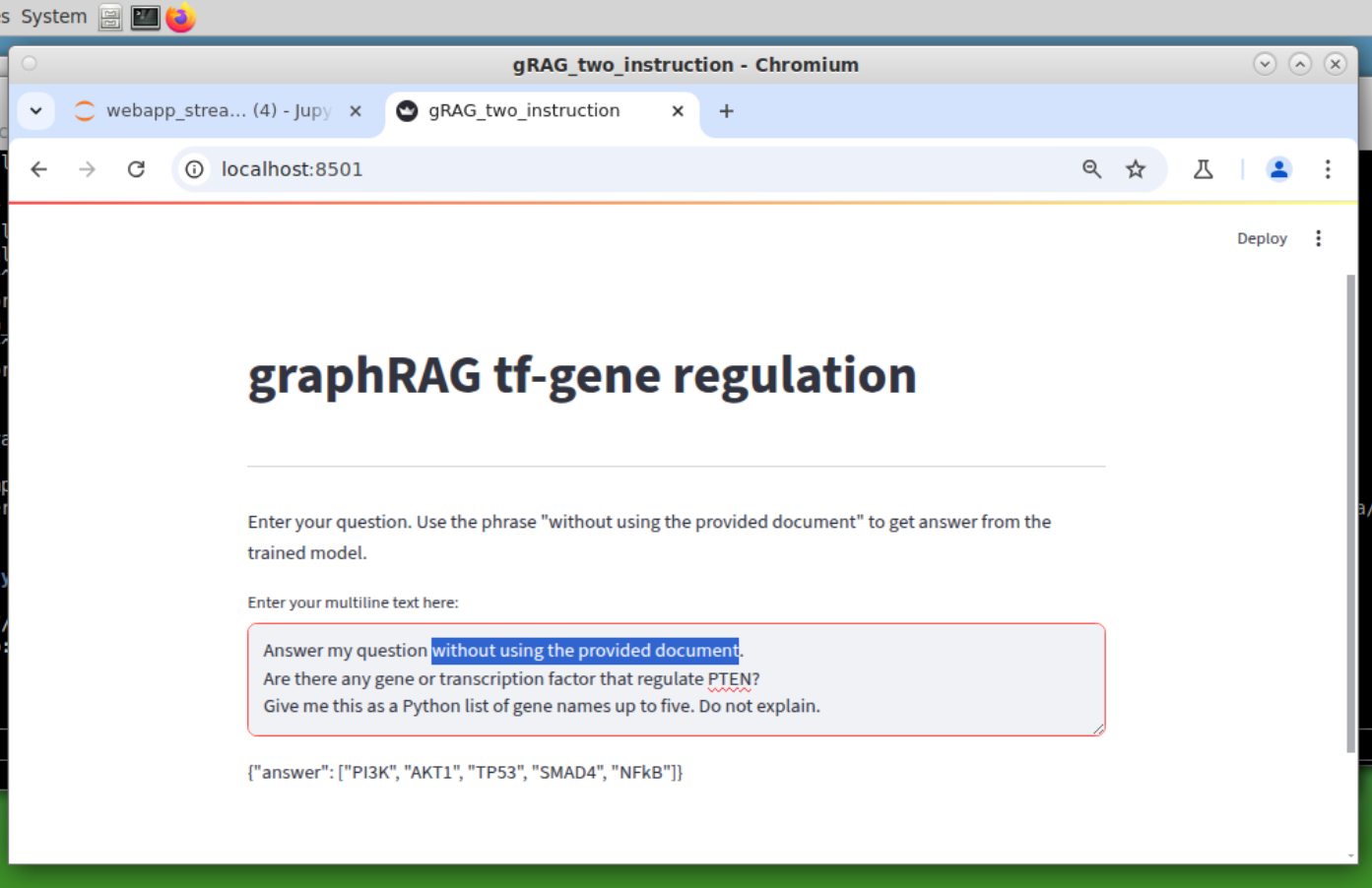
Two instruction version. Now the graphRAG can switch mode: with or without the graph DB.
Added context version. Now the graphRAG can add things that is not in the graph DB.